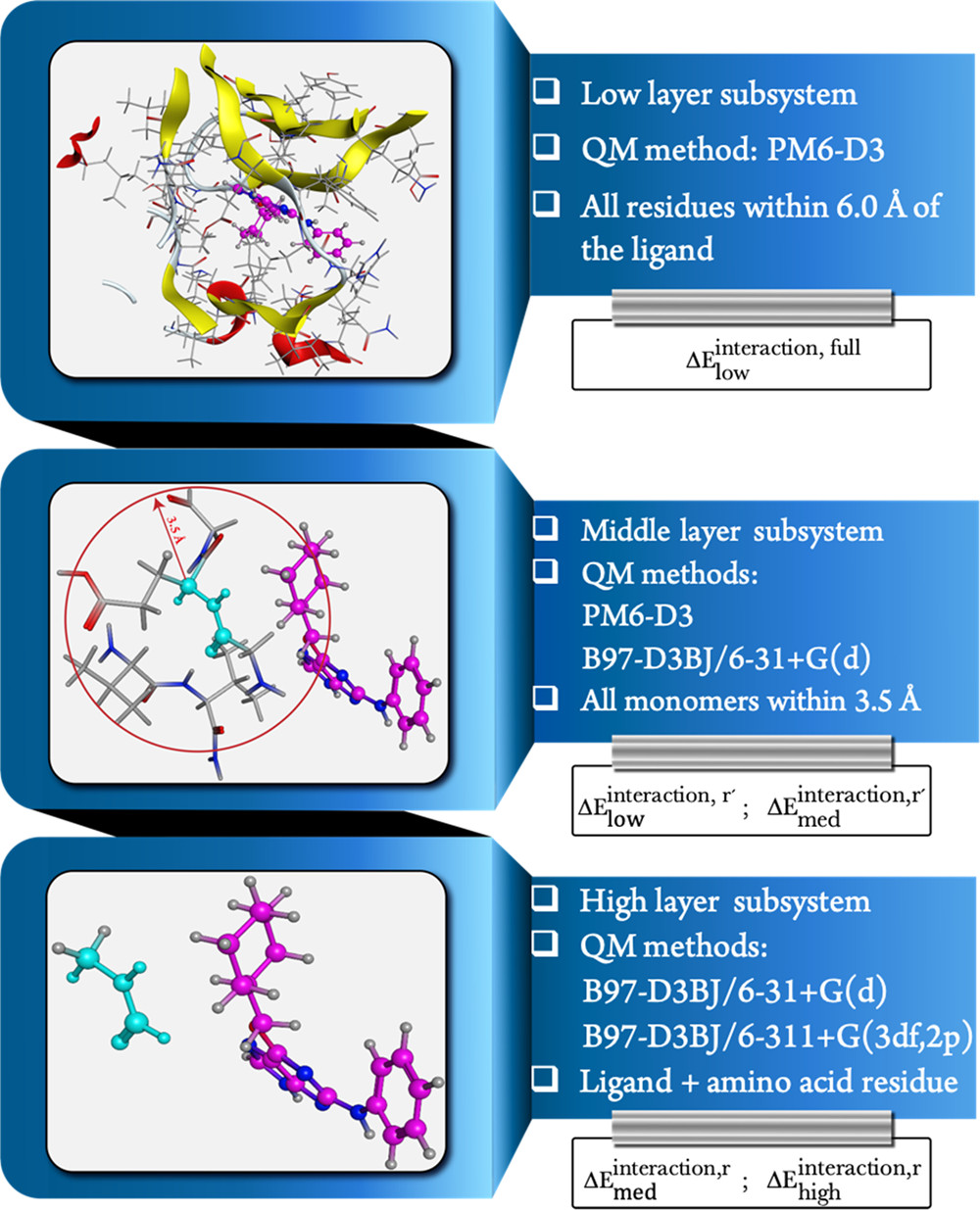
Using our molecules in molecules (MIM) fragmentation scheme, computationally efficient QM calculations can be used on large complex molecules (ex. proteins) to obtain accurate binding energies.
Recent Publications:
“Towards Post-Hartree-Fock Accuracy for Protein-Ligand Affinities Using the Molecules-in-Molecules Fragmentation-Based Method”, A. Gupta, S. Maier, B. Thapa, and K Raghavachari, J. Chem. Theory Comput. (2024). https://ddoi/10.1021/acs.jctc.3c01293
“Comparative Assessment of QM-based and MM-based Models for Prediction of Protein-Ligand Binding Affinity Trends”, S. Maier, B. Thapa, J. Erickson and K. Raghavachari, Phys. Chem. Chem. Phys. 24, 14525–14537 (2022). https://doi.org/10.1039/d2cp00464j
""Energy Decomposition Analysis of Protein-Ligand Interactions Using Molecules-in-Molecules Fragmentation-Based Method”, B. Thapa and K. Raghavachari, J. Chem. Inf. Mod. 59, 3474-3484 (2019 ). https://doi.org/10.1021/acs.jcim.9b00432